Software
R packages
Visit our GitHub site: github.com/odisce
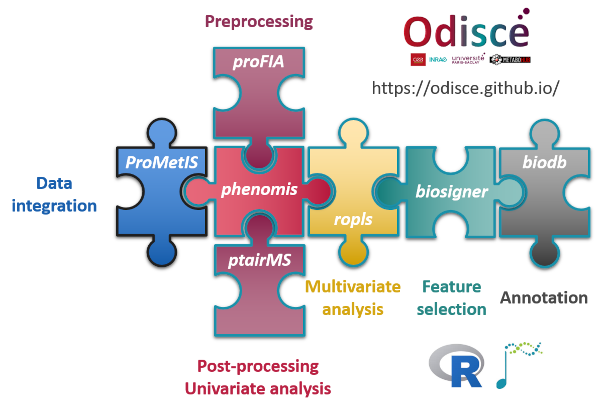
Preprocessing
proFIA: preprocessing of FIA-HRMS data (DOI:10.18129/B9.bioc.proFIA)
ptairMS: Pre-processing PTR-TOF-MS Data (DOI:10.18129/B9.bioc.ptairMS)
Spec2Xtract: Pre-processing of MS/MS raw spectra (https://github.com/odisce/Spec2Xtract)
DIANMF: processing of SWATH-DIA metabolomics data (https://github.com/odisce/DIANMF)
Post-processing
- phenomis: post-processing and univariate analysis of MS data (DOI:10.18129/B9.bioc.phenomis)
Machine learning
ropls: PCA, PLS(-DA) and OPLS(-DA) for multivariate analysis and feature selection of omics data (DOI:10.18129/B9.bioc.ropls)
biosigner: signature discovery from omics data (DOI:10.18129/B9.bioc.biosigner)
ProMetIS: multi-omics phenotyping of the LAT and MX2 knockout mice (https://github.com/IFB-ElixirFr/ProMetIS)
Cheminformatics
mineMS2: Annotation of spectral libraries with exact fragmentation patterns (https://github.com/odisce/mineMS2)
biodb: a library and a development framework for connecting to chemical and biological databases (DOI:10.18129/B9.bioc.biodb)
Datasets; MetaboLights repository
MTBLS1903: ProMetIS: deep phenotyping of knock-out mice by proteomics and metabolomics.
MTBLS404: Analysis of the human adult urinary metabolome variations with age, body mass index and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses (‘Sacurine’ data set).
MTBLS148: Impact of collection conditions on the metabolite content of human urine samples as analyzed by liquid chromatography coupled to mass spectrometry and nuclear magnetic resonance spectroscopy.
Galaxy modules; Galaxy toolshed
Batch Correction (in collaboration with Jean-Francois Martin, Melanie Petera, Marion Landi and Franck Giacomoni)
Biosigner
Heatmap
Multivariate
proFIA
Quality Metrics (in collaboration with Melanie Petera)
Univariate (in collaboration with Marie Tremblay-Franco)
Workflows and histories; Workflow4Metabolomics e-infrastructure
W4M00001_Sacurine-statistics (DOI:10.15454/1.4811121736910142E12)
W4M00002_Sacurine-comprehensive (DOI:10.15454/1.481114233733302E12)
W4M00003_Diaplasma (DOI:10.15454/1.4811165052113186E12)